Beginning with the discovery of penicillin by Alexander Fleming in the late 1920s, antibiotics have revolutionized the field of medicine. They have saved millions of lives each year, and have even been used prophylactically for the prevention of infectious diseases. However, we have now reached a crisis where many antibiotics are no longer effective. Such infections often result in an increased number of hospitalizations, more treatment failures and the persistence of drug-resistant pathogens.
Better understanding of mechanisms of antibiotic resistance would allow the development of control strategies to reduce the spread of resistant bacteria and their evolution. Bacteria may be intrinsically resistant to a class of antibiotics or may acquire resistance.
Main mechanisms of resistance to antibiotics can be caused by:
Alteration of the target site of the antibiotic. A common strategy for bacteria to develop antimicrobial resistance is to avoid the action of the antibiotic by interfering with their target site. To achieve this, bacteria have evolved different tactics, including protection of the target (avoiding the antibiotic to reach its binding site) and modifications of the target site that result in decreased affinity for the antibiotic molecule.
Enzyme inactivation of the antibiotic. One of the most successful bacterial strategies to cope with the presence of antibiotics is to produce enzymes that inactivate the drug by adding specific chemical moieties to the compound or that destroy the molecule itself, rendering the antibiotic unable to interact with its target.
Active transport of the antibiotic out of the bacterial cell. The production of complex bacterial machineries capable to extrude a toxic compound out of the cell can also result in antimicrobial resistance. Many classes of efflux pumps have been characterized in both Gram-negative and Gram-positive pathogens.
Decreased permeability of the bacterial cell wall to the antibiotic. Many of the antibiotics used in clinical practice have intracellular bacterial targets or, in case of Gram-negative bacteria, located in the cytoplasmic membrane (the inner membrane). Therefore, the compound must penetrate the outer and/or cytoplasmic membrane in order to exert its antimicrobial effect. Bacteria have developed mechanisms to prevent the antibiotic from reaching its intracellular or periplasmic target by decreasing the uptake of the antimicrobial molecule.
Main mechanisms of resistance to antibiotics can be caused

Bacteria can develop resistance to antibiotics by mutating existing genes (vertical evolution), or by acquiring new genes from other strains or species (horizontal gene transfer).
Many of the antibiotic mobile resistance genes (MGEs) are carried on genetic elements (plasmids, transposons or phages) that act as vectors that transfer these genes to other members of the same bacterial species, as well as to bacteria in another genus or species.
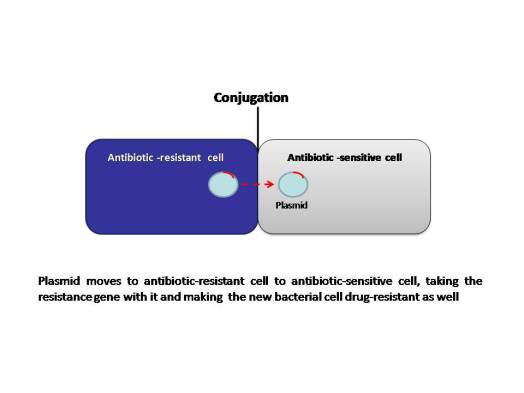
The MGEs allow resistance to spread horizontally and disseminate among different bacterial species. Although this association seems improbable, it appears to occur frequently and follows a series of evolutionary steps fueled by natural selection (antibiotic selection). The power of modern DNA sequence analysis allows us to better understand the process of emergence of these genetic structures. The primary mechanisms of horizontal transfer are conjugation (plasmids are transferred from a donor cell to a recipient cell), transformation (uptake of naked DNA), and transduction (bacteriophages as transporters of genetic information). Conjugation is considered the principal mode for antibiotic resistance transfer, because many resistance genes are situated on mobile elements, such as plasmids integrons and transposons.
Most families of antibiotics present in nature are compounds produced by fungi or bacteria; bacteria utilize these compounds to eliminate competitor microorganisms. As part of this arms race, many microorganisms code for genes whose products neutralize antibiotics; these genes may have been present in bacterial chromosomes for millions of years and they were probably not mobile, as evidenced by recent findings. The massive use of antibiotics probably favored selection of antibiotic resistant bacteria resulting in large numbers of bacteria coding for resistance genes.
On the other hand, bacterial chromosomes are populated with transposable elements (insertion sequences known as ISs), which jump frequently and randomly, as demonstrated during in vitro experiments from one genetic location to another in the same cell. Antibiotic resistance genes could be mobilized to genetic structures, such as plasmids and phages, which can move horizontally between bacterial cells including different bacterial species. The association of resistance genes to these mobile structures could occur through ISs; this has been postulated as the origin of many MGEs. Alternatively, plasmids or phages may also integrate in the bacterial chromosome in the vicinity of resistance genes and then mobilize the resistance genes as these structures excise from chromosomes.
Emergence of resistance among the most important bacterial pathogens is recognized as a major public health threat affecting humans worldwide. Widespread use of antibiotics has undoubtedly facilitated the development of antimicrobial resistance worldwide. Better understanding of the mechanisms of antibiotic resistance, will help clinicians regarding usage of antibiotics in different situations.
